This feature is available for republication under a Creative Commons licence.
In his new state-of-the-art microbiology lab, Professor Serge Mostowy manoeuvres what might be the world’s smallest syringe towards tiny zebrafish embryos.
“There” he says, indicating the microscope lens, his enthusiasm infectious “You can see the bacteria spreading, you can see the immune system responding. It’s right there in front of your eyes.”
A walk down the corridor takes us to the source of these embryos. It’s more like an aquarium than a lab, with hundreds of small tanks full of adult zebrafish. But a closer look shows this is no tourist attraction. Tanks labelled with terms such as “CD41 + GFP”, show how each group of fish has undergone gene editing to allow the study of different biological responses to infection.
“We’re just interested in the embryos” says Prof Mostowy, who leads a research group at the London School of Hygiene & Tropical Medicine (LSHTM). “We leave the adult fish to swim around, eat and mate.” A team of animal technicians keeps them fed, and each morning embryos are collected for study. “We have to get in quickly to collect them early during their development”, he adds.
As we return, Prof Mostowy points to a locked door, “that’s our new laboratory. Some of the pathogens we study are seen as dangerous in the wrong hands, so we need to meet strict rules to study them.” After a pause, he adds “But then, they can be also dangerous in nature, so we need to study them, and there aren’t many places in the UK where I can do this work.”
The use of zebrafish is an emerging approach to studying infection, and one Prof Mostowy has helped to pioneer. “In the first days of a zebrafish’s life, it is completely translucent” he explains excitedly. “So, we can actually see under a microscope what biological processes are happening in a live environment. You can’t do that in a mouse because it has skin and fur, but you can in a zebrafish.”
“This gives us phenomenal insight into what’s actually happening at a biological level when infection enters our body. And thanks to advances in microscopy” – he gestures to an impressive looking piece of kit labelled ‘Zeiss laser-scanning-confocal-microscope with Airyscan’ – “we can actually observe how pathogens interact with cells at a molecular level. If we understand the mechanisms different infections use to attack our cells and multiply, that’s the first step to finding targeted ways to stop them.”
This approach - of studying the fundamental biology of infection and developing responses aimed at individual pathogens - is attracting growing interest from global health programmes. Overuse of antimicrobials has led pathogens – bacteria, viruses and parasites - to evolve resistance. Resistance to antibiotics – the group of antimicrobials used to treat bacterial and some parasitic infections – is a particularly pressing problem, and antibiotics can also have unwelcome side-effects. Antimicrobial resistance (AMR) is considered by the World Health Organization to be one of the biggest threats to global health we are currently facing. If antimicrobials - our main weapon in the battle against infectious disease - become ineffective, we will face a health crisis not seen since the dawn of modern medicine.
“Evolution is a powerful force and we need to start thinking outside the box to stay ahead of the game,” says Prof Mostowy. “Often that means going back to fundamental biology to understand exactly what’s happening when our body becomes infected, so we can develop more creative ways to disrupt the pathogen’s ability to infect. Fortunately, recent advances in technology are allowing us to do just that”.
Are we winning the war on infection?
“Infection refers to a disease-causing agent that can be ‘caught’, for example from another human, an animal or the environment,” explains Professor Jimmy Whitworth, Professor of International Public Health at LSHTM. “It is the process by which pathogens invade your body”.
Prof Whitworth describes our two main weapons in the fight against these pathogens. Firstly, vaccines can be used to create immunity, preventing people acquiring an infection in the first place. Vaccines introduce a neutralised version of a specific infection which stimulate the body’s natural defences, creating an acquired immunity without actually contracting the disease. They are a major weapon in disease control.
Secondly, drugs and therapeutics allow us to respond to infection once it takes hold. Pathogens attack the body’s cells like an invading army. If the body can’t cope on its own, we can send in reinforcements, in the form of antimicrobials, which either destroy the pathogen directly or cut their supply lines, so they can’t reproduce.
“We need to start looking at responses targeted at individual pathogens, rather than a one-pill-fits-all approach”
Dr Lisa Dawson, Assistant Professor at LSHTM
For the last 70 years, antimicrobials have been our main therapeutic weapon against most bacterial infections including many chest, gut and urinary tract infections, and some parasitic ones such as malaria or the sexually transmitted infection, Trichomonas. In recent years antiviral drugs have also been developed which inhibit multiplication and transmission of viruses such as swine flu and HIV.
“Thanks to improving treatments, as well as public health measures, infectious diseases are falling. Globally they account for around 20% of disease, according to the Global Burden of Disease Study 2017, and in most places infectious disease has fallen below non-infectious conditions such as cancer and heart disease,” says Prof Whitworth.
This is cause for celebration but not complacency. By their nature, infectious diseases spread quickly if they are not contained. Until they are controlled, there is an ongoing threat that they will re-emerge. And that threat is growing.
Like any invading army, if we keep using the same defence strategy, the invaders will start to find ways around it. New strains of antimicrobial resistant pathogens are appearing all the time, creating an ‘arms race’ between pathogens and drug developers.
“To solve these challenges, we need to start looking at responses targeted at individual pathogens, rather than a one-pill-fits-all approach”, says Dr Lisa Dawson, an Assistant Professor at LSHTM whose research is also tackling this issue.
Now is a good time for such an approach, claims Michael Chew, Science Portfolio Adviser, Infection & Immunobiology at the Wellcome Trust, which funds health research at institutions around the world, including LSHTM. “The latest genome sequencing and bioinformatics tools are now very accessible, increasing our ability to study pathogens at the molecular level. Gene sequencing and editing allow researchers to manipulate the precise mechanisms by which pathogens attack human cells. New microscopes and animal models allow the infection itself to be followed in real time.”
Using bacteria to fight bacteria
In the west, diarrhoea is often seen as inconvenience after an undercooked meal. It often surprises people to hear that it is the world’s second biggest killer of children under-five.
Shigella bacteria is the second leading cause of deaths from diarrhoea worldwide (after Rotavirus). It is responsible for approximately 165 million cases of disease and kills around 160,000 people globally per year, according to a 2018 Lancet article.
Like its famous cousin, E. coli, Shigella attacks the human gut. It is easily transmitted, and young children are particularly susceptible. Most infections occur in poor communities, striking down children living in densely populated areas, where sanitation is lacking, and there is little access to the clean drinking water they need to counter life-threatening dehydration.
Although Shigella infection (called shigellosis) can be treated with antibiotics, there are an increasing number of drug resistant strains. Shigella is now one of the 'superbugs’ deemed a priority by the World Health Organization.
Back in Prof Mostowy’s lab, these Shigella bacteria are whisked down the length of a syringe and injected into zebrafish embryos for analysis.
“We follow the bacteria’s progress and watch exactly how the zebrafish’s immune system responds to infection,” he says. “We can get an immediate handle on the mechanisms by which a bug causes disease and start thinking about how to treat it.”
“Prof Mostowy’s approach is very novel”, says Michael Chew. “The zebrafish model, cutting edge imaging and tagging with dyes, allow in vivo studies of how the host tries to stop and get rid of invasive pathogens.”
In one piece of research, a collaboration with Prof Liz Sockett at the University of Nottingham, zebrafish were used to test a treatment using Bdellovibrio; predatory bacteria which inserts itself into other bacteria and destroys them from within.
In the study, Bdellovibrio was labelled red and Shigella green, by engineering bacteria to express fluorophores so they can be clearly observed in the zebrafish. Using their high-resolution microscopes, the team saw that Bdellovibrio actively killed the Shigella bacteria and rescued zebrafish from Shigella infection. Perhaps more amazingly, Bdellovibrio was most efficient in clearing Shigella when it worked together with the natural zebrafish immune system.
This is important not just for Shigella, but as a potential approach to combat many other antibiotic resistant organisms. Bdellovibrio uses hundreds of enzymes to degrade bacteria, so it’s hard for the bacteria they attack to develop resistance. There are several other predatory bacteria which act in a similar way, so there is a lot of research to be pursued in this area, and zebrafish are the ideal starting point for testing the potential of each in a living organism.
“We are looking beyond how to destroy entire populations of bacteria, as antibiotics research often does,” says Prof Mostowy. “Bacteria can be viewed as individuals. We need to spot the 1% of bacteria within populations that don’t respond to antibiotics and understand how else they can be targeted. We are looking at biological processes of antibiotic resistance, and fundamental concepts of immunity and host defence.”
Harnessing the body’s natural defences
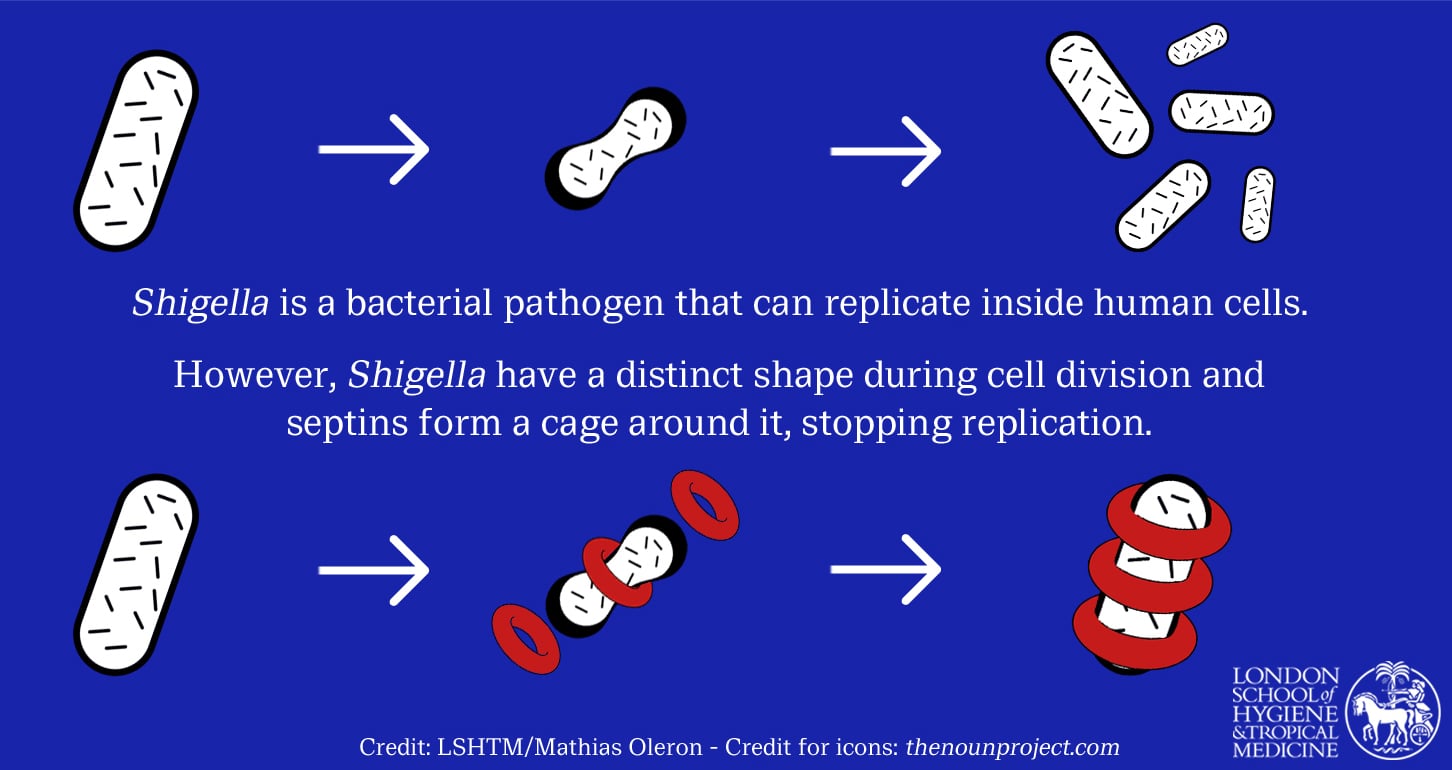
Prof Mostowy is also interested in how the human body’s own defences can be harnessed to fight bacteria such as Shigella, perhaps with a little nudge. Another success for his team came from research into septins, proteins in human cells which recognise bacteria and form a cage around them to stop them from replicating.
Recent research established the mechanism by which septin cages work. Prof Mostowy’s team found that septins recognise the physical shape of Shigella bacterium where it divides. Remarkably, when septins spot this specific geometry, they form a cage-like structure around it.
“This biological language is an ancient evolutionary process, but it is one we are only just beginning to understand. This is a natural host cell ‘sensor’ recognising a shape, which indicates dividing bacterial cells, and responding to it to protect the host. Because this is a response to fundamental bacterial properties, it works regardless of antimicrobial resistance.”
Future work is required to determine how septins can be harnessed for therapeutic purposes, but it is hoped that novel drugs can be developed to boost septins in the body, thereby promoting a natural defence mechanism. In 2013, the team discovered septin cages are conserved in humans and zebrafish, meaning the zebrafish model could be used in future studies to fully investigate bacteria which septins recognise.
Some say zebrafish are too dissimilar to humans to be useful, but Prof Mostowy disagrees. “Sure, there is a leap before we can develop human treatments, but we view the zebrafish as a discovery platform. We can employ a wide variety of approaches in the zebrafish and discover what’s happening at a biological level. If promising, we can move to mice and eventually clinical trials.”
His confidence seems well placed. Following the zebrafish research, Bdellovibrio is now being tested in mice and human blood. If successful, this could become a new tool for site-specific infections in patients – for example for wounds or surgery – in the not-too-distant-future.
Shigella: the "superbug"
Where did all the good bacteria go?
The human gut contains a diverse range of ‘good bacteria’ called the microbiome, which ensures healthy digestion. Unfortunately, widespread antibiotic use and changing diets has wreaked havoc on the delicate ecosystem of human guts, depleting good bacteria and leaving many people with digestion problems and reduced immunity to infection.
Enter Clostridium difficile (C. diff). These opportunistic bacteria exist naturally in some people’s gut, and in most cases the gut’s microbiome keeps it in check. Until the 1970s it rarely caused much of a stir, and before 2004 surveillance of C. diff cases was not mandatory. But when good bacteria are depleted, usually as a result of antibiotic usage, bad bacteria such as C. diff can take hold. Once it does, it releases toxins that damage gut cells, causing severe diarrhoea. In 2017 C. diff caused over 13,000 infections in the UK, with 64% acquired in the community and 14% in hospital. In the USA up to half a million people suffer from C. diff infections yearly, with around 15,000 deaths (2015).
This creates a Catch-22 situation. Antibiotics can get rid of C. diff, but in doing so will further destroy good bacteria, making it harder to control C. diff and other bad bacteria in future.
What’s more, C. diff produces antibiotic resistant spores that can sit quietly in the gut until the antibiotics are gone, then grow back, returning infection in around 35% of cases. Of particular worry is that C. diff, once confined to the over 65s, is becoming more common in younger people as the bacteria evolves.
LSHTM’s Dr Lisa Dawson is studying C. diff. Armed with the genetic code of the bacteria, which she and her team recently unravelled, Dr Dawson is looking at new ways to tackle C. diff without antibiotics.
“We start by asking how we can we target these bacteria specifically. Do they have certain novel traits, and can we develop inhibitors to block these? What do they need to survive, and can we prevent them accessing that?” she says.
Hidden in the genetic code was a useful discovery. C. diff produces a nasty compound called para-cresol which kills healthy bacteria. Para-cresol is also used in bleach and sewage treatment. In a weakened gut, C. diff can reach a critical mass where it has the upper hand, and can then use para-cresol to stop the gut’s good bacteria from returning.
The team experimented with removing the gene that produces para-cresol and studied the effect on the microbiome of mice. They discovered that good bacteria could establish themselves much quicker when para-cresol was neutralised, and there were fewer relapses.
This insight prompted Dr Dawson to start looking for chemicals that inhibit the gene which creates para-cresol. “We are looking at ways to interfere with a bacteria’s defence mechanisms, allowing the microbiome to recover and the body to do its job of supressing C. diff”, she says. It is hoped this will lead to a drug that will stop production of para-cresol, which could reduce the need for antibiotics.
“Many scientists now agree that the natural bacteria in the gut have an important role in health, and Dr Dawson’s approach of helping good bacteria tackle bad bacteria is gaining popularity,” says Chew.
Septins restrict Shigella cell division
Halting a new malarial outbreak in South East Asia
Fighting infection is a game of cat and mouse. Pathogens evolve defences against our weapons as fast as we can develop them. Changes in our body’s biology opens new ways in for opportunistic pathogens. But it’s not just the biological goalposts that keep moving, sometimes changes to our physical world are behind outbreaks of new diseases.
For many years, Malaysia has been cutting down its forests to make room for profitable crops such as palm oil. As humans have moved in to farm these new crops, they have come into close contact with macaque monkeys, which previously lived deep in the rainforests away from human settlements.
This led to an unusual form of malaria parasite called Plasmodium knowlesi starting to infect humans. The closer contact has meant that mosquitoes could easily move between humans and monkeys, now living side-by-side. The number of human cases of so called ‘monkey malaria’ has increased rapidly, up tenfold since 2008.
Such diseases are known as zoonoses, pathogens transmitted from animals to humans. They are a cause of many dangerous human infections, including Ebola and bird-flu, often appearing following changing land use and disruption of animal ecosystems.
Dr Rob Moon, Assistant Professor of Molecular Parasitology, runs a research group at LSHTM from his specialist malaria lab, recently completed as part of the same modernisation project that created a home for Prof Mostowy’s zebrafish. Alongside international partners, Dr Moon is hoping to stop P. knowlesi in its tracks.
His work takes advantage of recent innovations in gene sequencing and gene editing, in particular CRISPR-Cas9, which has made gene editing relatively simple. Using this technique, researchers can send in ‘molecular scissors’ which cut a section of DNA so genes can be added or removed. This was previously very difficult to do with any precision. By observing the effect that removing different genes has on parasite behaviour, they can establish which genes play a role in infection.
His research has generated a complete picture of the changes to P. knowlesi’s genetic code that occur when it adapts to infect human cells. In doing so, the team discovered that the mechanism by which P. knowlesi gets into human red blood cells requires a gene called NBPXa. When they disabled this gene, the parasite could no longer get inside cells, and could not multiply.
They are now looking at ways to stimulate the body’s defences in a way which neutralises NBPXa. “If successful, this could pave the way for a vaccine against P. knowlesi malaria, preventing this emerging disease taking hold” says Dr Moon.
“We are studying how malaria infections occur, looking at the genes that allow pathogens to take hold and how we can manipulate these to stop the parasite in its tracks. Once we know what genes or processes are essential, we can develop ways to target them, in this case by triggering an antibody response that can block parasite invasion. Critically, we can now quickly test and optimise the effectiveness of these responses against cultured malaria parasites, ensuring only the best vaccine candidates get taken forward for expensive vaccine trials in animals or humans.”
“Eradication programmes using antimalarials, insecticides and bednets have been very effective in South East Asia,” Dr Moon adds. “Whilst this works well for other human malaria parasites, for P. knowlesi this is still a huge reservoir of parasites within monkeys, so the monkeys represent a real wild card for control. A vaccine is the real prize, as it could allow for a single intervention, after which we can walk away knowing the whole population is protected - no matter what is going on in the monkeys!”
The world has seen just one human disease, smallpox, eradicated globally since the dawn of modern medicine, as well as several at a regional level, including malaria in many parts of the world. Malaysia has the infrastructure to coordinate a vaccination programme which would eradicate human P. knowlesi in the region. They just need the right biological tools.
Infection in the real world
Understanding the biology behind infection is the start, but not the end, of the story. Research needs to be turned into practical and affordable treatments, which must be delivered to people in diverse parts of the world. “This research into fundamental biological processes is the start of a chain of infection control measures which goes on to include observation, modelling, community studies, vaccination programmes, public health initiatives, and community outreach” says Prof Whitworth.
In the labs of Mostowy, Dawson and Moon, and others like them around the world, the innovative science being carried out will underpin real-world infection control measures in years to come. As we face changes in modes of infection - whether from evolving pathogens, changes to our physiology, or exposure to new strains of disease – such research offers hope of a new line of defence.
“New tools and ways of working, such as new animal models to improve our understanding of the immune system, genomic sequencing, and CRISPR-Cas9 gene modification, make this an exciting time for infectious disease research”, concludes Chew. “They also highlight the direction of future research and the importance of funding these innovative approaches.”
“These threats are clearly a huge and daunting challenge for humankind” Prof Mostowy adds. “But it is a great time to be a researcher; there is a real appetite right now around the world for new, creative ways to control infection.”
Photographic commission by James Sykes - follow on Instagram @jamessykesphoto
In making these images I wanted to replicate the methodical, scientific approach of the practitioners who inhabit this space, and move away from the oft repeated images of science in action. I utilised an approach called ‘dead pan’ that has a rich history in contemporary photography, a formal approach with maximum depth of field ensuring everything is in focus with the intention of democratising the components of the image rather than presenting them hierarchically. The situation is depicted neutrally, without comment. However there are human elements to many of the pictures, the tweezers labelled with a name, the pen waiting to be written with, lab coats waiting to be worn. The portraits are intended to echo this almost sterile approach and focus on the humans as subjects. There is a separation between the people and the science, intended to highlight the relationship between, neither aspect having significance without the other.
Mostowy lab photo gallery: Image 1 - stereo fluorescent microscope (Leica M205-FA), used to image fluorescent zebrafish transgenic and/or zebrafish infected with fluorescent bacteria; image 2 - Zeiss LSM 880 confocal microscope with Airyscan; image 3 - confocal microscopy suite; image 4 – electroporator; image 5 - stage controller and power supply; Zeiss AxioObserver Z1 for live-cell fluorescence imaging; image 6 – a set of pipettors in the tissue culture hood; image 7 - petri dishes carrying injected zebrafish larvae, researchers can maintain 6-100 larvae in a single dish; image 8 – long-finned zebrafish; image 9 - bench of a zebrafish researcher, bacteria from larvae can be plated on agar plates to count colony forming units, single larvae are loaded in 24 well plates to monitor infection over time; image 10 - tweezers (used to manually hatch zebrafish eggs) and eppendorfs containing reagents (for lysis of larvae for CFU quantification); image 11 - Nagisa Yoshida; image 12 - Hoan Ngo, PhD, Royal Society – Newton International Fellow; image 13 - Vincenzo Torraca, postdoctoral researcher; image 14 – (l-r) Hoan Ngo, Ana Lopez Jimenez, Stevens Robertin, Nagisa Yoshida, Vincenzo Torraca, Margarida C. Gomes, Serge Mostowy. Credit: James Sykes.
We hope you enjoyed reading about our work in this feature. If you are interested in supporting projects like these and the many others we are leading to improve health worldwide, we would be delighted to hear from you. There are many ways you can make a gift to the School, from wherever you are in the world. Each and every gift we receive makes an impact, from funding scholarships, to updating our facilities or investing in new avenues of research. Whether it’s a gift of £5 or £500,000 your generosity will support our mission to improve health in the UK and around the world.