
Exterior of LSHTM building
Johnson and Johnson Satellite Centre for Global Health Discovery at LSHTM
The Johnson and Johnson Satellite Centre for Global Health Discovery at LSHTM is focused on advancing the development of drug regimens for the treatment of tuberculosis to counter the threat from AMR.
The Satellite Centre for Global Health Discovery, based at the London School of Hygiene & Tropical Medicine (LSHTM), aims to develop exploratory biology for new drugs to combat tuberculosis (TB). This deadly disease was responsible for 1.6 million deaths in 2021 and accounts for nearly a third of all AMR-related deaths. The centre aims to support the development of new therapeutics to improve drug treatment options for TB, achieved in part by supporting active drug discovery biology programmes.
Research at the centre is focused on advancing current drug discovery efforts as well as providing a better understanding of the biology surrounding key drug targets in TB. This includes studying the mode of action of new and known drugs in order to improve their effectiveness and reduce the risk of resistance. The centre utilises a matrix of techniques to achieve its goals including transcriptomic, metabolomic and genomic approaches combined with classical in vitro assays.
Recent updates
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory
Events
Newsletter
Contact us
Prof Anil Koul
Anil.Koul@lshtm.ac.uk
Department of Infection Biology
Dr Richard Wall
Richard.Wall@lshtm.ac.uk
Department of Infection Biology

The Johnson & Johnson Satellite (J&J) Centre for Global Health Discovery (CGHD), based at the London School of Hygiene & Tropical Medicine (LSHTM), aims to develop new drugs to combat tuberculosis (TB). The centre belongs to a global network of research partnerships that combine leading academic research institutes with J&J's expertise in drug discovery for tuberculosis to expedite early-stage research and development needed to address global threats to health. The CGHD at LSHTM will specifically support the development of new therapeutics to improve drug treatment options for TB.
TB is an infectious disease caused by a bacterium called Mycobacterium tuberculosis. TB was responsible for 1.6 million deaths in 2021 and almost a third of all deaths related to drugs. Bedaquiline, belonging to a new class of TB drugs called diarylquinolines, is a medication used to treat drug-resistant TB. It was discovered by scientists at J&J and is the only new anti-TB drug with a novel mode of action to be approved for the treatment of drug resistant TB in nearly 50 years. It is primarily used as a second-line treatment for patients who have drug-resistant TB.
The Satellite Centre for Global Health Discovery at LSHTM aims to continue and expand on the success of bedaquiline by using cutting-edge techniques to develop the next generation of anti-TB drugs including metabolomics or other omics approaches for new target exploration. Combining the best of industry and academic environments will accelerate lifesaving innovations from the lab to the clinic.
Recent updates
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory
Events
Newsletter
Contact us
Prof Anil Koul
Anil.Koul@lshtm.ac.uk
Department of Infection Biology
Dr Richard Wall
Richard.Wall@lshtm.ac.uk
Department of Infection Biology

Anil
Koul
Professor of Translational Discovery

Richard
Wall
Assistant Professor
John
Dallow
Research Assistant
Ellie Davis
Scientific Officer
Cora Dee
Project coordinator

William
Pearson
Research Fellow
Recent updates
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory
Events
Newsletter
Contact us
Prof Anil Koul
Anil.Koul@lshtm.ac.uk
Department of Infection Biology
Dr Richard Wall
Richard.Wall@lshtm.ac.uk
Department of Infection Biology
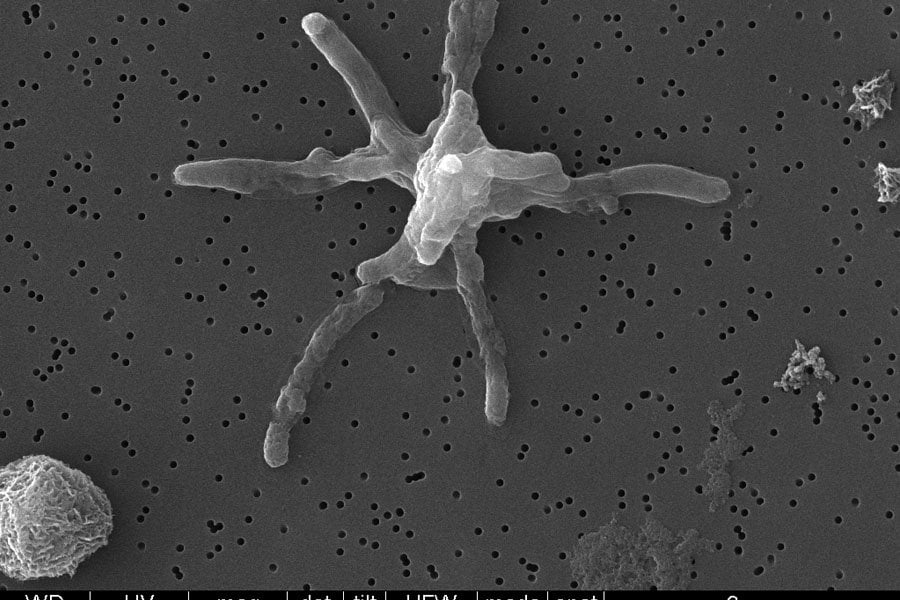
Tuberculosis (TB) is an infectious disease caused by the bacterium Mycobacterium tuberculosis. In 2021, TB was responsible for 1.6 million deaths and almost a third of all deaths related to antimicrobial resistance (AMR). New treatments can improve treatment outcomes, shorten the duration of treatment and reduce the risk of drug resistance, making it an important step in controlling and eventually eliminating the disease. At the Centre for Global Health Discovery at LSHTM, we are focused on understanding the biology surrounding key drug targets in TB, identifying new starting points for drug development and advancing our understanding of TB drug mode of action and resistance. To achieve these goals, we focus on the following areas of TB drug development research related to key hotspots in TB metabolic pathways:
Investigation of key metabolic pathways
Metabolomics is the study of metabolic pathways and their corresponding small molecule products within a biological system. In TB, metabolomics can help to understand and explore drug targets by providing a comprehensive view of the metabolic changes that occur within the bacterium. By analysing the metabolic profiles of bacteria under different conditions, such as in the presence or absence of drugs, we can identify new targets for drug development. For example, the identification of essential metabolic pathways can lead to the discovery of enzymes or other metabolic components that are required for the survival of the bacterium, and that can be targeted by drugs to disrupt its growth and replication.
Identification of key genes involved in drug action and resistance including drug efflux
RNA sequencing (RNA-seq) is a powerful tool for the analysis of gene expression in biological systems. It can help to understand and explore drug targets by providing a comprehensive view of the changes in gene expression that occur in bacteria in response to drug treatment. By sequencing the RNA from bacteria treated with different drugs, we can identify changes in gene expression that are associated with drug efficacy and toxicity. This information can be used to identify new targets for drug development. Additionally, RNA-seq can also be used to monitor changes in gene expression over time during drug treatment, which can provide insight into the dynamics of drug action and the evolution of drug resistance both in vitro and preclinical models.
Characterisation of in vitro drug action
The pathology of TB infection is complicated with bacteria residing in very different conditions within the patient. In vitro experiments that manipulate environmental factors such as varying the energy source or oxygen levels can help to replicate these conditions allowing us to better predict how a drug will behave in these different conditions. These experiments can also produce insight into the metabolic pathways and processes that are essential for the survival of the bacteria.
Drug target identification of phenotypically active compounds
Drug target deconvolution is an important aspect of TB drug discovery and development. We use genomic, proteomic and transcriptomic approaches to deconvolve drug targets of phenotypically active compounds in TB. For example, resistant line generation can provide valuable information for understanding and exploring drug targets. When a bacterial strain develops resistance to a particular drug, it is often due to a change in the genetic makeup of the bacteria, such as a mutation in a specific gene. By studying the genetic changes that occur in the resistant strains, we can identify the genes involved in resistance and explore the possibility of targeting these genes as new drug targets. Additionally, by creating resistant strains, we can test new treatments and evaluate their efficacy against the resistant bacteria. These different techniques are instrumental in advancing our understanding of TB biology and identify potential drug targets that will provide starting points for new drug discovery programmes.
Elucidation of drug mechanism of action
We are using techniques to modulate protein expression to understand the biology surrounding key drug targets in TB. For example, CRISPR-interference (CRISPRi) can be used to study the role of specific genes related to drug action by reducing the expression of genes of interest. Studies using CRISPRi have been used previously to identify potential drug targets for TB by altering the activity of specific genes and observing the effects on the growth and survival of TB bacteria. These studies have been used to identify genes that are essential for the survival of TB bacteria, making them potential targets for new drugs. Additionally, CRISPRi has been used to identify genetic variations in TB strains that may contribute to resistance, which can inform the development of new drugs that target these specific variations. Conversely, overexpression of enzymes and proteins involved in drug action, as well as the drug target, allows us to study their function and validate them as potential drug targets.
We are always interested in collaborating with other researchers including drug target deconvolution work with phenotypically active compounds. Please get in touch with Richard Wall to discuss future collaborations.
Recent updates
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory
Events
Newsletter
Contact us
Prof Anil Koul
Anil.Koul@lshtm.ac.uk
Department of Infection Biology
Dr Richard Wall
Richard.Wall@lshtm.ac.uk
Department of Infection Biology
Navigating the diversity of Mycobacterium tuberculosis populations found in clinical settings can be challenging, especially when trying to correlate resistance-associated mutations identified through target deconvolution studies. To bridge this gap, we've developed an accessible online database and made this easily accessible to the scientific community via an open-access dashboard. It compiles 1,063,811 non-synonymous genetic variations across 694,579 sites from a vast collection of 51,183 clinical TB isolates, covering the entire protein-encoding genome of M. tuberculosis. This open-access resource serves as an invaluable tool for exploring the prevalence of resistance mutations in clinical contexts.
To use this dashboard in your own work please use the following reference:
Phelan, J, Van den Heede, K, Masyn, S, Verbeeck, R, Lamprecht, DA, Koul A, Wall, RJ (2024) An open-access dashboard to interrogate the genetic diversity of Mycobacterium tuberculosis clinical isolates. Scientific Reports. 14: 24792.
For more information, contact Jody Phelan or Richard Wall.
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory Medicine at the University of Cape Town and Professor of Mycobacteriology and Global Health at LSHTM, who discussed the impending threat of bedaquiline-resistant tuberculosis in South Africa and the steps required to minimise its spread.
Read the full interview.
We are delighted to share our recent work on the compiling the genetic diversity of M. tuberculosis clinical isolates. The database, made available via an open-access dashboard, serves as an invaluable tool for exploring the prevalence of resistance mutations in clinical contexts. Read more about this in our recent paper published in Scientific Reports.
We are pleased to release our latest work: an accessible online database of genetic variance from 51,183 clinical TB isolates, made available to the scientific community via an open-access dashboard. Covering the entire protein-encoding genome of M. tuberculosis, this database serves as an invaluable tool for researchers exploring the prevalence of resistance mutations in clinical contexts.
The dashboard is designed to be user-friendly, enabling researchers to seamlessly integrate this data into their own work. This open-access resource provides the scientific community with additional tools to better understand and address resistance-associated mutations in clinical settings.
Richard has recently joined the Antimicrobial Resistance (AMR) centre management committee as a committee member.
In this role, he intends to assist in the hosting of more drug discovery-related events at LSHTM as well as explore opportunities to teach the subject of AMR via school outreach programmes in the near future.
Presentation by Professor Sir Stewart Cole KCMG FRS 'From TB drug development to the Ineos Oxford Institute for antimicrobial research'.
For those who missed it, we're happy to share the video recording of the seminar.
Chaired by Prof Anil Koul and hosted by the TB Group at LSHTM, this was held in the John Snow A lecture theatre on 14 March, 2024.

We were delighted to host Professor Sir Stewart Cole at LSHTM this week where he delivered a fantastic talk on the history of TB drug discovery, the current pipeline and future perspectives. He also discussed his time at the Institut Pasteur and his latest role at the Ineos Oxford Institute for antimicrobial research (IOI).

Ellie together with Tesnim and Terry Kipkorir recently hosted 4 applied science college (pre-university) students for a weeklong work experience in collaboration with The Switch, a charity based in Tower Hamlets. They provide students from disadvantaged backgrounds with opportunities to learn essential skills and form contacts that may not be otherwise available to them. Providing them with such valuable experiences not only enriches their understanding of science but also equips them with practical skills and accomplishments to highlight on their CVs.
William joins us as a Research Fellow following time at Imperial College London. He will predominantly work on TB metabolomics in the group.
John and Richard recently had the pleasure to meet with teams from the other two J&J Centres for Global Health Discovery, based at NUS-Duke (Singapore) and H3D (South Africa). Richard was able to present the important contributions that the LSHTM team have had in J&J's TB drug discovery portfolio.

We welcome Ms Tesnim Alattar and Ms Ellie Davis who join the group as Scientific Officers!

Dr Sherif Abouelhadid and Richard had the pleasure of interviewing Prof Koul where they discussed the threat of antimicrobial resistance, the challenges of working with tuberculosis bacteria and how the Centre is helping to expedite TB drug discovery to find the next generation of treatments. Read more from their conversation.
Recent updates
As part of World Antimicrobial resistance Awareness Week (WAAW), Richard interviewed Prof Keertan Dheda, Professor of Respiratory
Events
Newsletter
Contact us
Prof Anil Koul
Anil.Koul@lshtm.ac.uk
Department of Infection Biology
Dr Richard Wall
Richard.Wall@lshtm.ac.uk
Department of Infection Biology