Dissecting the Red Blood Cell Invasion Pathways of the Malaria Parasite Plasmodium knowlesi
Location of study: LSHTM
LSHTM Investigators: Robert W. Moon, Franziska Mohring, Melissa Hart, James Charleston, Colin Sutherland, Susana Campino
External collaborators: Simon Draper (Jenner Institute, UK), Helen Saibil (Birkbeck, UK), Mike Blackman (Francis Crick Institute, UK), Tony Holder (Francis Crick Institute, UK), Arnab Pain (King Abdullah University of Science and Technology, Saudi Arabia), Jake Baum (Imperial College London, UK, Neil Almond (National Institute of Biological Standards and Control, UK), Eun Taek Han (Kwandon National University, Korea)
Funding Body: Medical Research Council and Department for International Development through an MRC Career Development Award to Rob Moon., Bloomsbury Research Studentship to Melissa Hart, BBSRC LIDO Studentship to James Charleston
Malaria is caused by single-celled parasites called Plasmodium. Symptoms of malaria arise from the parasite destroying red blood cells (RBCs) by entering them, multiplying within and bursting out again in a continuous cycle. The parasites produce a range of adhesive proteins enabling them to bind to the surface of RBCs and invade them. These parasite proteins can determine disease severity and which hosts are susceptible, as well as presenting important targets for vaccine design.
This MRC Career Development award Fellowship, jointly funded by the MRC and DfID, investigates the role of these adhesive proteins during RBC invasion using a malaria parasite known as Plasmodium knowlesi. This naturally infects macaques in SE Asia, and is now known to cause of severe and fatal human infections. We have adapted CRISPR Cas9 genome editing techniques to P. knowlesi and used this to systematically tag and knock out each of the genes with putative roles in invasion, determining that two, NBPXa and DBPa are required for invasion of human red blood cells. Using live cell imaging and electron tomography we will now determine their precise role during invasion. We have also successfully developed new tools to allow us to use P. knowlesi to aid vaccine development for other important malaria parasites like P. vivax, which remain difficult to study within the lab.
Identification of a potent series of inhibitors of the malaria parasite cGMPdependent protein kinase that clear blood stage infection in vivo and block transmission
Location of study: LSHTM and LifeArc (formerly MRC Technology)
LSHTM Investigators: David Baker, Lindsay Stewart, Win Gutteridge, Simon Croft, Hans Dessens, Teun Bousema and Eloise Walker.
External collaborators: Simon Osborne, Katy Kettleborough, Andy Merritt, Jon Large, Kristian Birchall, Keith Ansell and colleagues, (LifeArc, UK); Javier Gamo, Laura Sanz and colleagues, (GSK, Spain); Ray Hui and colleagues, (Structural Genomics Consortium, University of Toronto, Canada); Koen Dechering, (TropIQ Health Sciences, The Netherlands)
Funding Body: MRC Developmental Pathway Funding Scheme
Due to the emergence of drug resistant malaria parasites new antimalarial drugs must be developed urgently. We have generated a highly potent series of inhibitors of the cGMP-dependent protein kinase (PKG) that clear blood stage infection in an in vivo P. falciparum model and block transmission of gametocytes to mosquitoes.
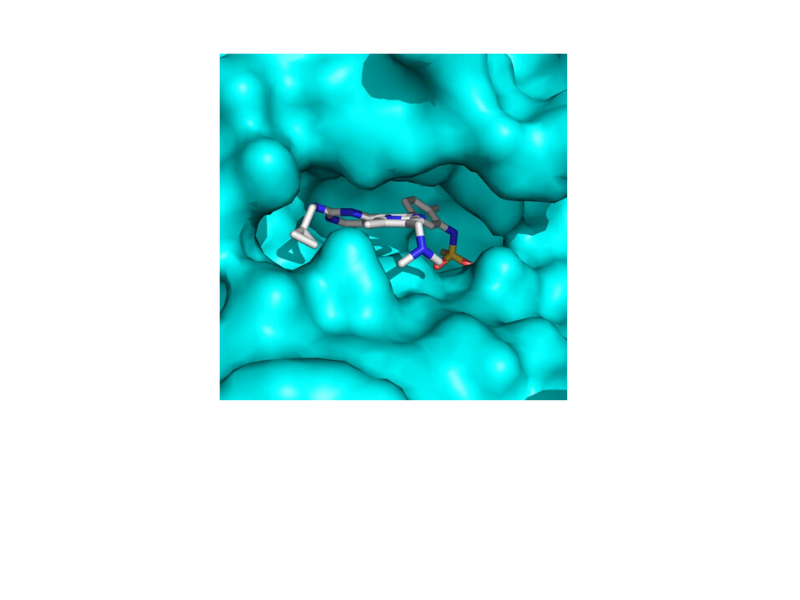
The malaria parasite cGMP-dependent protein kinase (PKG) is essential for all the key stages of the complex malaria parasite life cycle and so a drug that targets this enzyme has the potential to interrupt the malaria life cycle at multiple stages. Through partnership with LifeArc (formerly MRC Technology), we carried out a medicinal chemistry programme and developed a highly potent imidazopyridine series of PKG inhibitors. The best compound (ML10) blocks blood stage Plasmodium falciparum development with an EC50 value of ~2 nM. Importantly, the series also has potent activity against sexual stages and blocks transmission to mosquitoes. Using a chemical genetic approach we showed that ML10 is highly selective for the malaria parasite PKG. ML10 cleared blood stage parasitaemia in a GSK in vivo Plasmodium falciparum model providing proof of concept for PKG as an antimalarial drug target. Finally, through partnership with scientists at the Structural Genomics Consortium, we were able to determine the co-crystal structure of PKG bound to ML10 revealing intimate molecular contacts that explain the high levels of potency and selectivity we have observed. The structural data will greatly assist future medicinal chemistry efforts to optimise the series.
Multi-population genomic analysis of malaria parasites indicates local selection and differentiation at the gdv1 locus regulating sexual development
Location of study: LSHTM UK, Gambia, Guinea, Senegal, Mali, Mauritania and Ghana
LSHTM Investigators: Craig W. Duffy, Alfred AmambuaNgwa, Sarah J. Tarr, Lee Murray, Lindsay B. Stewart, Umberto D’Alessandro, David J. Conway
External collaborators: Ambroise D. Ahouidi (UCAD, Senegal), Mahamadou Diakite (University of Bamako, Mali), Gordon A. Awandare (WACCBIP, University of Ghana), Hampate Ba (INRSP, Mauritania), Thomas D. Otto, Dominic P. Kwiatkowski (Wellcome Sanger Institute)
Funding Body: European Research Council (ERC), Medical Research Council (MRC) and Royal Society
Parasites infect hosts in widely varying environments, encountering diverse challenges for adaptation. Identifying the genes under local selection in different environments can help reveal potential targets for interventions.
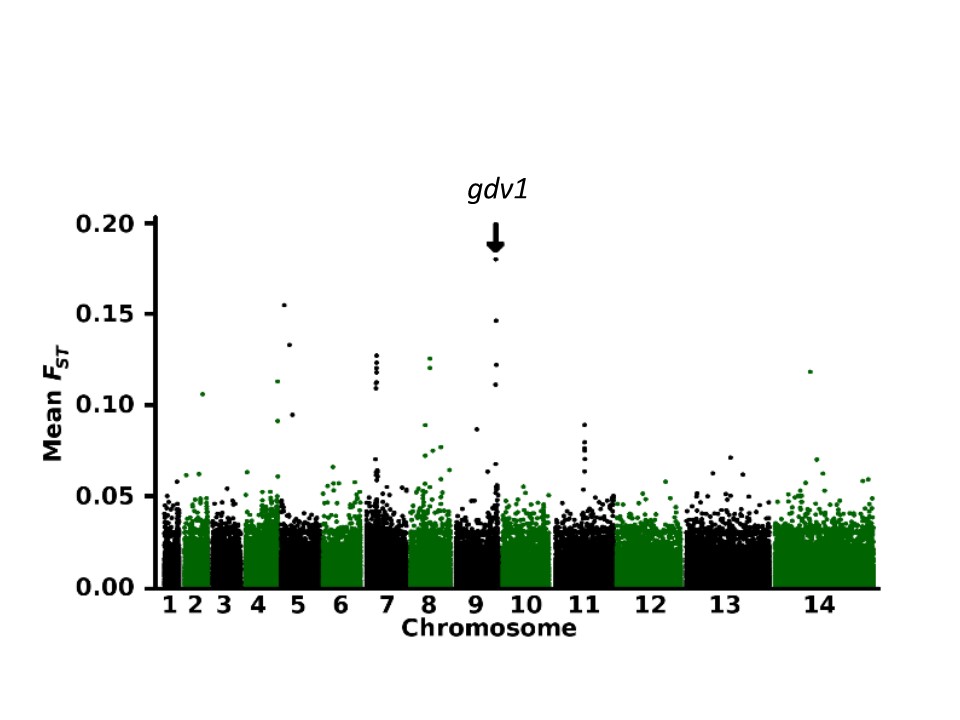
To identify malaria parasite genes under locally divergent selection across a large endemic region with a wide spectrum of transmission intensity, genome sequences were obtained from 284 clinical Plasmodium falciparum infections from four newly sampled locations in Senegal, The Gambia, Mali and Guinea. Combining these with previous data from seven other sites in West Africa enabled a multi-population analysis to identify discrete loci under varying local selection. A genome-wide scan showed the most exceptional geographical divergence to be at the early gametocyte gene locus gdv1 which is essential for parasite sexual development and transmission.
We identified a major structural dimorphism with alternative 1.5 kb and 1.0 kb sequence deletions at different positions of the 3’-intergenic region, in tight linkage disequilibrium with the most highly differentiated single nucleotide polymorphism, one of the alleles being very frequent in Senegal and The Gambia but rare in the other locations. Long non-coding RNA transcripts were previously shown to include the entire antisense of the gdv1 coding sequence and the portion of the intergenic region with allelic deletions, suggesting adaptive regulation of parasite sexual development and transmission in response to local conditions.